Project View
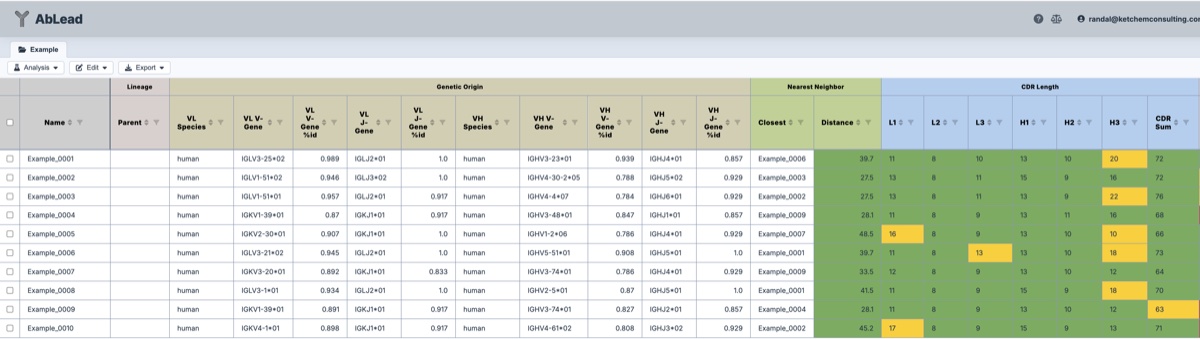
The Project View provides a summary of all antibodies in the project.
- Sort & Filter: Click a header sort button to sort or its filter button to enter a filter value. Filtering is context sensitive and can filter by text or value ranges. Click the filter button again to clear a filter. Click the sort button again to sort in the opposite direction.
- Name: The entry name associated with the analysis. Names may be double-clicked to be edited.
- Lineage: The parent of the entry. If the entry is a variant, the parent is the antibody it was derived from. These may be set by the user, but are automatically set of Engineering builds.
- Genetic Origin: The genetic origin and species of the antibody chains. This is determined as the closest germline sequence. These values are helpful for determining genetic diversity, comparison to known germline stabilities (but that can change with a single mutation), and known germline titer (but that can change with a single mutation).
- Nearest Neighbor: The closest sequence to that entry within the set. This is a simple depiction of set diversity. Clading provides a more detailed view of set diversity.
- CDR Length: The measured sequence lengths, as well as the summed total, for the CDRs based on the defined region scheme.
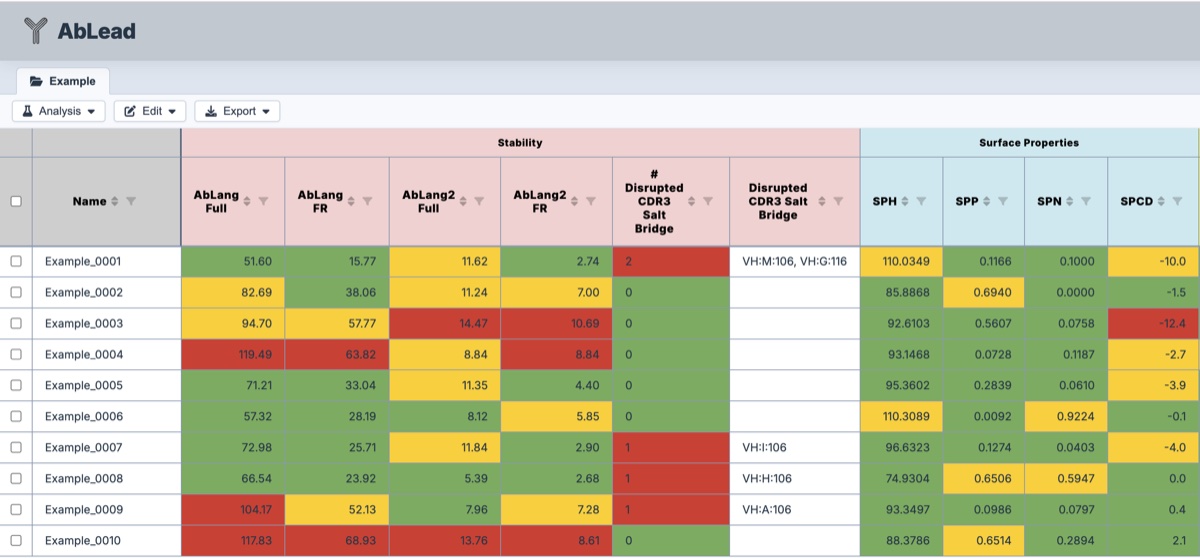
- Stability
- AbLang and AbLang2: Summed LLM likelihood differences between the parent and most likely residues at each position for full-length sequence and framework positions only. AbLang is a broader LLM while AbLang2 removes germline bias. See AbLang and AbLang2.
- Disrupted CDR3 Salt Bridge: Identifies if the canonical HC-CDR3 salt bridge has been disrupted by mutation. This can impact fold stability.
- Surface Properties
These are calculated similar to the OPIG TAP method.- SPH: Hydrophobicity character of the surface.
- SPP: Positive charge character of the surface.
- SPN: Negative charge character of the surface.
- SPCD: Fv charge interaction character of the surface.
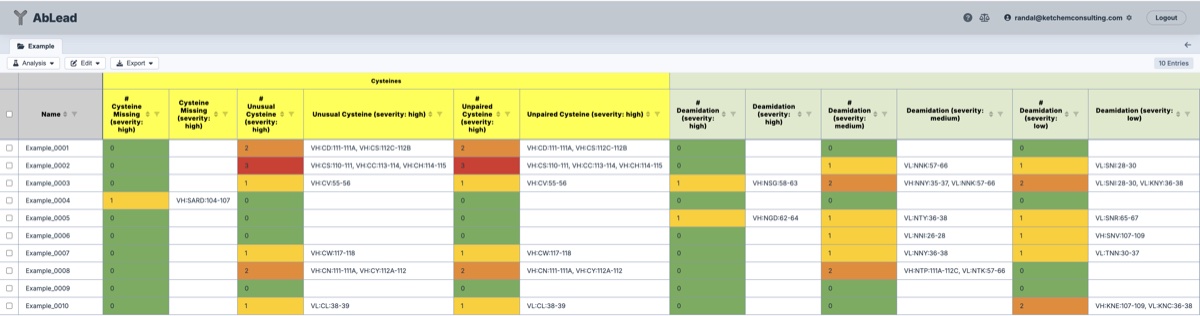
- Cysteines
- Cysteine Missing: Identifies if a normal Ig fold cysteine is missing from the sequence.
- Unusual Cysteine: Identifies cysteines in the sequence that are not in the normal Ig fold positions.
- Unpaired Cysteine: Identifies unpaired cysteines in the sequence. After structure evaluation these may be reclassified as paired in the Excel export to lower the score (lower is better).
- Potential PTMs
See LAP: Liability Antibody Profiler by sequence & structural mapping of natural and therapeutic antibodies.- Deamidation and Isomerization: If these are realized over time they could degrade antigen binding.
- N-linked Glycosylation: Identifies potential glycosylation sites. While these could help solubilize the Fv, they may also impact affinity and efficacy if there are differences across productions.
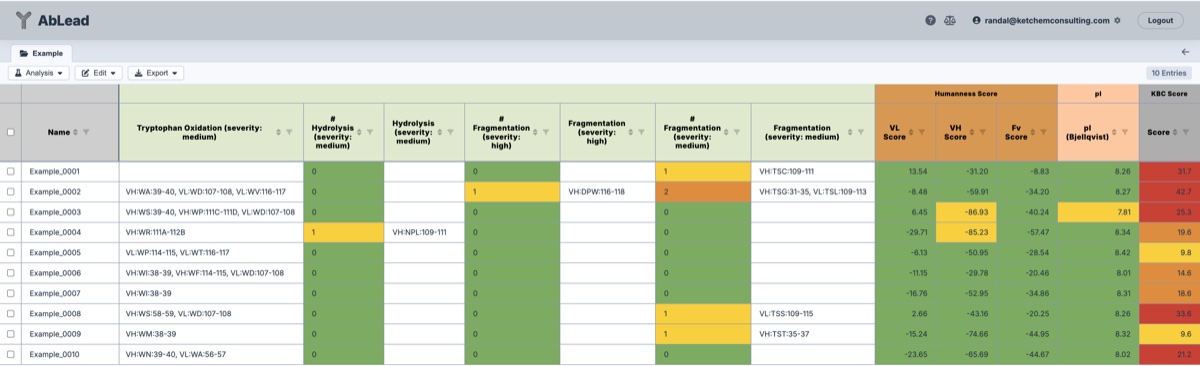
- Met and Trp Oxidation: Methionine and Tryptophan residues are susceptible to oxidation. If realized over time they could degrade antigen binding.
- Hydrolysis: Creation from NP to DP during deamidation, which could then isomerize.
- Fragmentation: Potential cleavage between defined residues.
- Humanness
A measure of the VL, VH, and Fv humanness of the sequence. AntPack calculates the log-probability of this sequence given its original training set of sequences. A higher score indicates a more human sequence. - Score
The score of the entry, calculated based on the analyses performed. This value may be used for rank ordering the set. The function used in the score calculation is exposed in the Excel output. The values and weights used for the KBC score may be modified in the user settings section, as well as in the Excel export. - Edit
- Add Fvs and/or PDBs: Add entries to the project and/or add PDBs to existing entries.
- Set Parent: Set or clear the parent of the selected entries.
- Delete Selected: Delete the selected entries. These are deleted from the project and the database immediately.
- Export
- Excel: Export the selected entries to an Excel file with conditional formatting and a weighted function to generate the scores.
The attributes used to generate the score, each with their own weight, are: L1, L2, L3, H1, H2, H3, CDR Sum, AbLang FR, Disrupted CDR3 Salt Bridge, SPH, SPP, SPN, SPCD, Cysteine Missing (severity: high), Unusual Cysteine (severity: high), Unpaired Cysteine (severity: high), Deamidation (severity: high), Deamidation (severity: medium), Deamidation (severity: low), Isomerization (severity: high), N-glycosylation (severity: high), Methionine Oxidation (severity: low), Tryptophan Oxidation (severity: medium), Hydrolysis (severity: medium), Fragmentation (severity: high), Fragmentation (severity: medium), Humanness Score: Fv Score, pI (Bjellqvist) - CSV: Export the selected entries to a CSV file.
- FASTA: Export the selected entries to a FASTA file.
- PDB Files: Export the stored PDB files for the selected entries.
- Excel: Export the selected entries to an Excel file with conditional formatting and a weighted function to generate the scores.

Genetic Origin, CDR Length, Potential PTMs, and Humanness are determined using AntPack.
References
- The Potential PTMs are described in Satława et al., PLoS Comp Biol (2024).
- Genetic Origin, CDR Length, Potential PTMs, and Humanness are determined using AntPack.
- AbLang details are described in Olsen et al., Bioinformatics Advances (2022).
- AbLang2 details are described in Olsen et al., bioRxiv (2024).
- The background inspiration for the Surface Properties (Tap-like method) is described in Raybould et al., PNAS (2019).