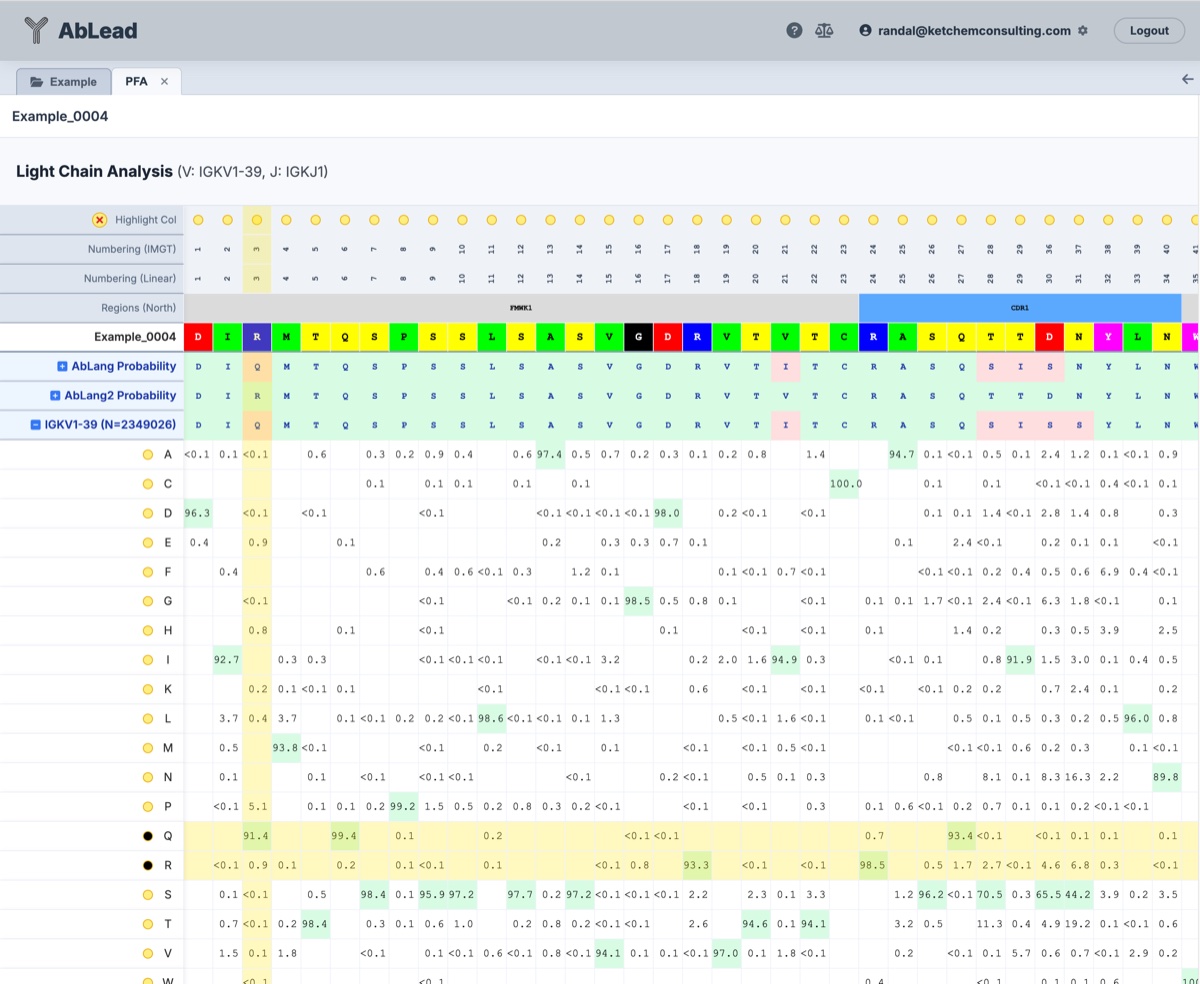
Position Frequency Analysis (PFA)
The PFA view provides a statistical and LLM-based breakdown of amino acid distributions at each position.
- Coloring: Highest frequency or likelihood residues are indicated by red highlighting of the sequence.
- Grid Exposure: Open a grid to visualize the probabilities for all residues at each position. Note that human germlines are searched regardless of parent species.
- Engineering Mutation Designs: Click a residue to enter a mutation design used by the Engineering tab.
- Observations: Click a residue to enter an observation for that IMGT position. These are displayed in the Observations tab and on a clicked residue.
The AntPack training set of 58,788,431 heavy chain and 68,454,444 light chain human sequences was used to calculate the PFA values.
The AbLang and AbLang2 grid values are generated by the models as residue likelihoods. A higher positive value indicates a higher likelihood of the residue at that position. The difference in the likelihood between the parental and highest likelihood residue indicates the severity of the difference, with large differences indicating a possibility that the parental position is not optimal and could be mutated to improve stability.
References
- AbLang details are described in Olsen et al., Bioinformatics Advances (2022).
- AbLang2 details are described in Olsen et al., bioRxiv (2024).